Application Examples:
1. Search applications:
(1) piRNA-centric application
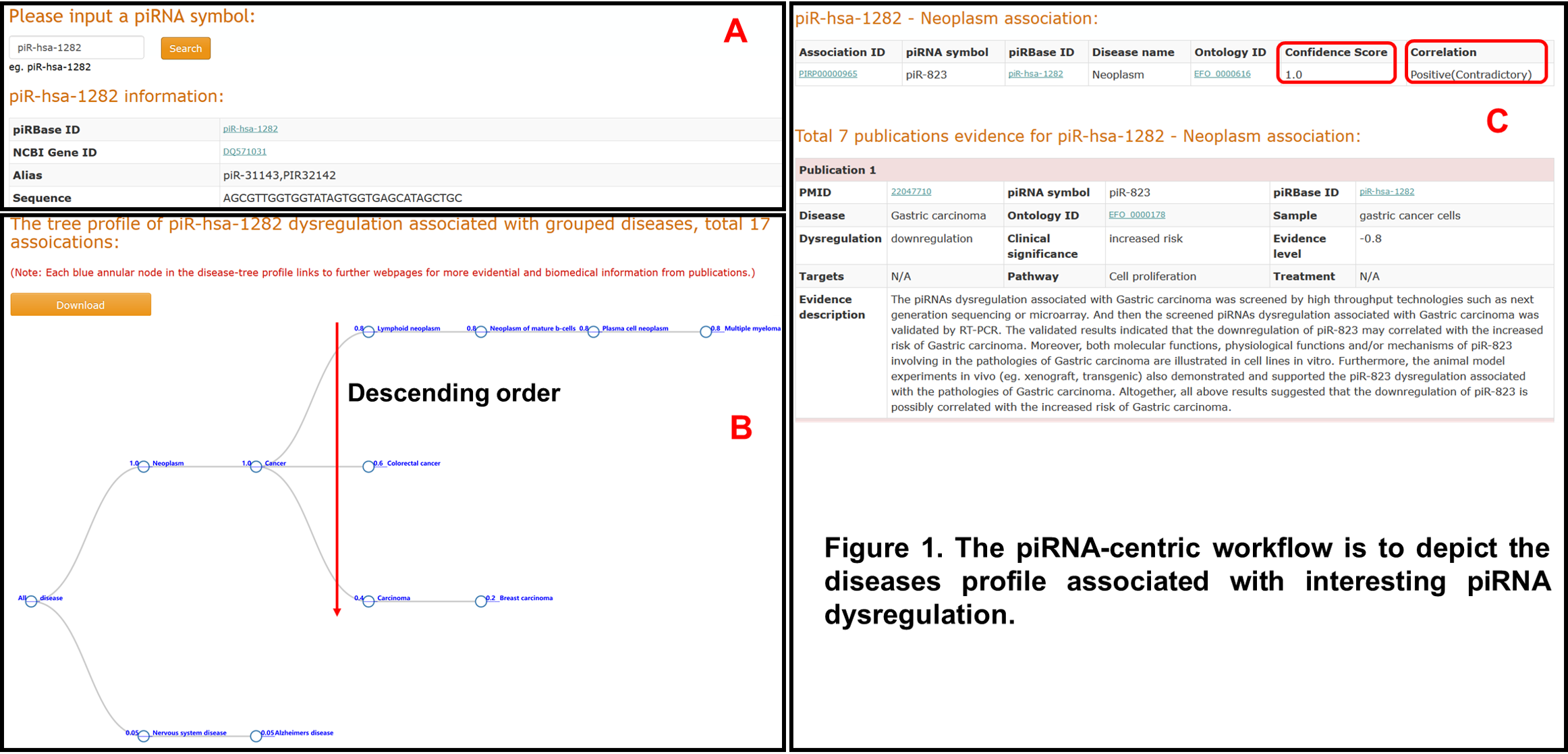
The piRNA-centric workflow is to depict the diseases profile associated with interesting piRNA dysregulation. Users can input an interesting piRNA in piRPheno to search and acquire a tabular profile for the piRNA information (Figure 1A) and a disease-tree visualization (Figure 1B). Here a disease-tree presents a group of diseases associated with the input piRNA in a tree structure with prioritization of confidence scores (Figure 1B). The diagram of disease-tree can be downloaded by clicking the “Download” button in the Figure 1B. Our research results suggest that the confidence score of 1 would be a highly reliable threshold to assess the confidence level of an association. Each blue annular node in the disease-tree profile (Figure 1B) links to other webpages for more information of the association, including Association_ID, correlation status, evidence levels, PMIDs, EFO terms, dysregulation, clinical significance, sample, pathway, treatment, target and evidence description of the association (Figure 1C). For example, the correlation of piR-823-neoplasm association is assigned with “Positive(Contradictory)” . This assigned means that there have conflicting evidences, which supports the piR-823 expression level positively associated with neoplasm. In addition, the PMID numbers and the EFO terms externally link to the databases of PubMed and EFO for more information (Figure 1C).
(2) Disease-centric application
The disease-centric workflow is to depict the piRNAs dysregulation profile associated with interesting disease. Users can input an interesting disease to search and acquire tabular profiles for the disease information (Figure 2A) and a prioritized piRNAs dysregulation associated with the input disease (Figure 2B). The tabular profile of piRNA-disease associations can be downloaded by clicking the “Download Table” button in the Figure 2B.The confidence score of 1.0 would be a highly reliable threshold to assess the confidence level of an association. Moreover, the prioritized associations profile is also visualized as a word-cloud diagram (Figure 2C) by clicking the ‘WordCloud visualization’ button (Figure 2B). The sizes and locations of the piRNA symbols in the word-cloud diagram represent the piRPheno confidence scores of the piRNA-disease associations (Figure 2C). Larger sizes and centralizations of piRNA symbols indicate higher confidence of the associations between the piRNAs and rheumatic disease in this case study (Figure 2. C). In addition, each “Association_ID” in the tabular profile (Figure 2B) links to other webpages for more information of the association, including Association_ID, correlation status, evidence levels, PMIDs, EFO terms, dysregulation, clinical significance, sample, pathway, treatment, target and evidence description of the association (Figure 2D).

2. Browse applications:
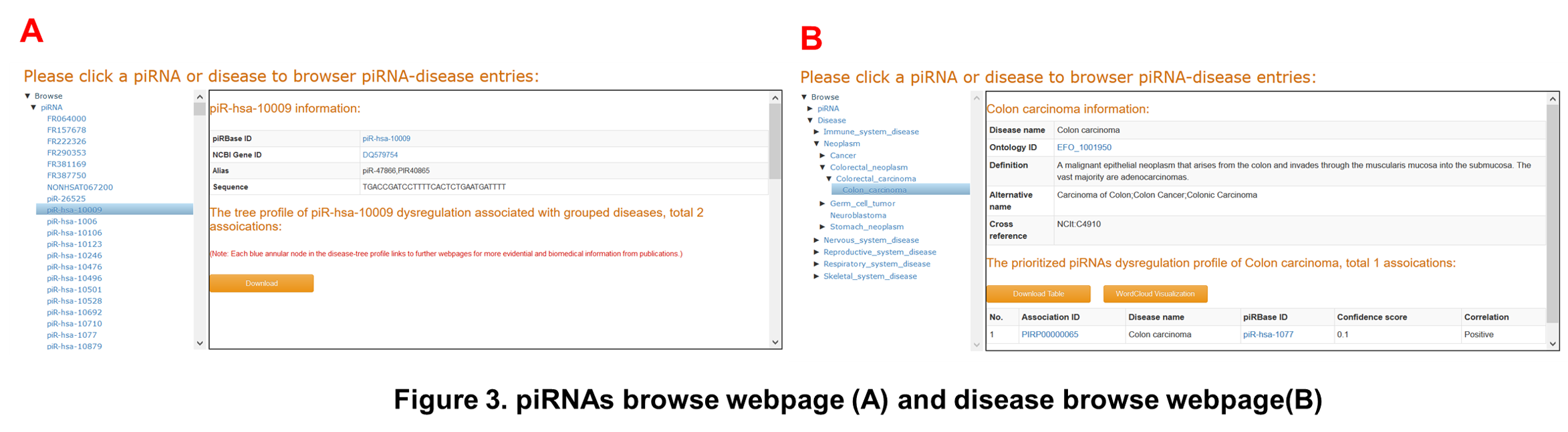
The piRNAs list and the diseases list in the tree structure are displayed in ‘Browse’ webpage (Figure 3). It allows users to easily browse interesting piRNAs (Figure 3A) and diseases (Figure 3B) for further information.
4. Web services
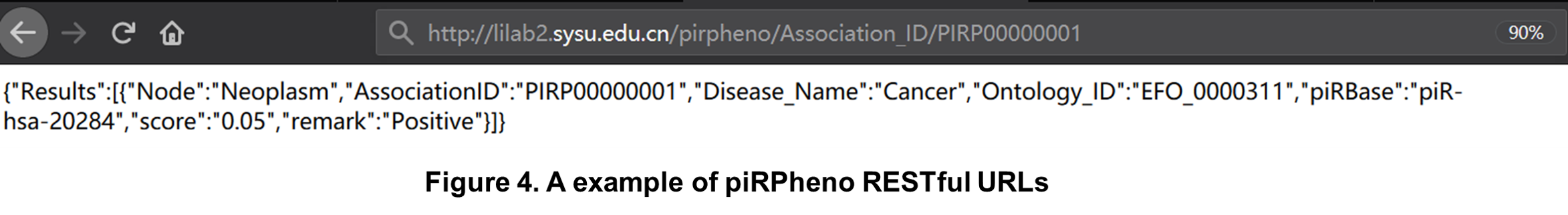
piRPheno RESTful URLs can be bookmarked, linked and used in programs for all entries, queries and tools available through this website. Data is available in universal JSON format (Figure 4) provided on the website for piRPheno association data. All resources are accessible using simple URLs (REST) that can be bookmarked, linked and used in programs. The simple URL address for an entry consists of a unique ‘Association_ID’ in ncRPheno. eg: http://lilab2.sysu.edu.cn/pirpheno/Association_ID/PIRP00000001.
5. Download
The Download webpage allows users to download all publication supported piRNA-disease association data in piRPheno, including piRNA-disease associations in “piRNA_Diseas_association.txt’’ file, piRNA related information in “piRNA_information.txt”, disease related information in “Disease_information.txt” file and the evidence information in “Evidence_information.txt” file. The data in ncRPheno is regularly updated; to download the latest dataset, please send a request to liweizhong@mail.sysu.edu.cn OR zhangwl25@mail2.sysu.edu.cn (Figure 5).

6. Submission
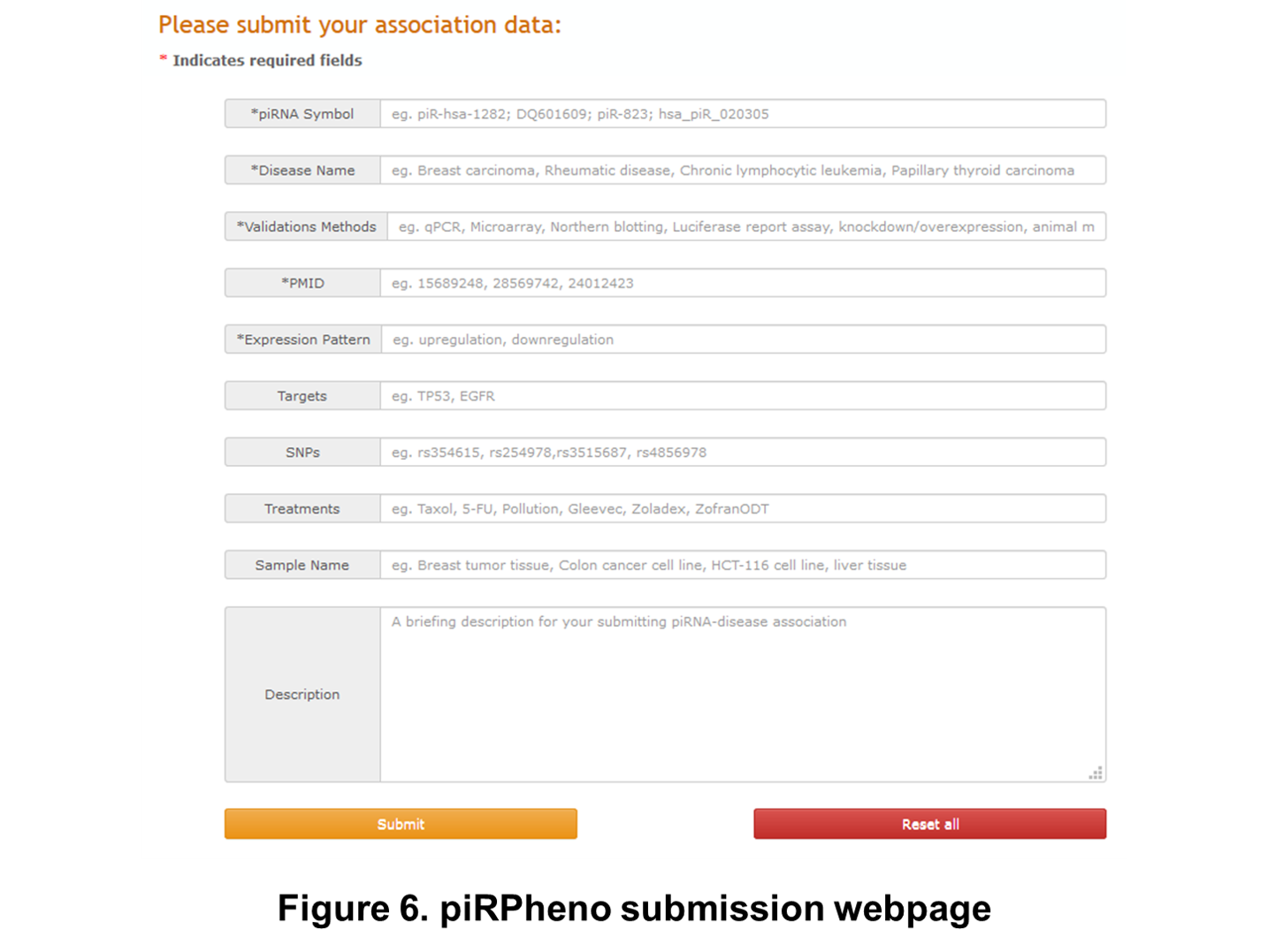
In the Submit webpage, piRPheno invites users to submit novel piRNA-disease association data. Users need to provide certain information that describes the candidate associations. Users are required to fill the textboxes containing a "*". Once approved by our submission review committee, the submitted association will be included in the piRPheno platform and made available to the public in the following release. (Figure 6).
7. Contact us
In case you experience any trouble using piRPheno or have any suggestions or comments, please do not hesitate to contacting us via e-mails: liweizhong@mail.sysu.edu.cn or zhangwl25@mail2.sysu.edu.cn